 View on TensorFlow.org View on TensorFlow.org
|
 Run in Google Colab Run in Google Colab
|
 View source on GitHub View source on GitHub
|
 Download notebook Download notebook
|
This tutorial provides examples of how to load pandas DataFrames into TensorFlow.
You will use a small heart disease dataset provided by the UCI Machine Learning Repository. There are several hundred rows in the CSV. Each row describes a patient, and each column describes an attribute. You will use this information to predict whether a patient has heart disease, which is a binary classification task.
Read data using pandas
import numpy as np
import pandas as pd
import tensorflow as tf
SHUFFLE_BUFFER = 500
BATCH_SIZE = 2
2024-08-16 06:59:37.935480: E external/local_xla/xla/stream_executor/cuda/cuda_fft.cc:485] Unable to register cuFFT factory: Attempting to register factory for plugin cuFFT when one has already been registered 2024-08-16 06:59:37.956364: E external/local_xla/xla/stream_executor/cuda/cuda_dnn.cc:8454] Unable to register cuDNN factory: Attempting to register factory for plugin cuDNN when one has already been registered 2024-08-16 06:59:37.962738: E external/local_xla/xla/stream_executor/cuda/cuda_blas.cc:1452] Unable to register cuBLAS factory: Attempting to register factory for plugin cuBLAS when one has already been registered
Download the CSV file containing the heart disease dataset:
csv_file = tf.keras.utils.get_file('heart.csv', 'https://storage.googleapis.com/download.tensorflow.org/data/heart.csv')
Downloading data from https://storage.googleapis.com/download.tensorflow.org/data/heart.csv 13273/13273 ━━━━━━━━━━━━━━━━━━━━ 0s 0us/step
Read the CSV file using pandas:
df = pd.read_csv(csv_file)
This is what the data looks like:
df.head()
df.dtypes
age int64 sex int64 cp int64 trestbps int64 chol int64 fbs int64 restecg int64 thalach int64 exang int64 oldpeak float64 slope int64 ca int64 thal object target int64 dtype: object
You will build models to predict the label contained in the target column.
target = df.pop('target')
A DataFrame as an array
If your data has a uniform datatype, or dtype, it's possible to use a pandas DataFrame anywhere you could use a NumPy array. This works because the pandas.DataFrame class supports the __array__ protocol, and TensorFlow's tf.convert_to_tensor function accepts objects that support the protocol.
Take the numeric features from the dataset (skip the categorical features for now):
numeric_feature_names = ['age', 'thalach', 'trestbps', 'chol', 'oldpeak']
numeric_features = df[numeric_feature_names]
numeric_features.head()
The DataFrame can be converted to a NumPy array using the DataFrame.values property or numpy.array(df). To convert it to a tensor, use tf.convert_to_tensor:
tf.convert_to_tensor(numeric_features)
WARNING: All log messages before absl::InitializeLog() is called are written to STDERR
I0000 00:00:1723791580.394635 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.398535 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.402304 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.406024 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.417906 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.423162 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.426583 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.430147 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.433654 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.437125 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.440588 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791580.443941 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.673556 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.675740 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.677839 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS ha
<tf.Tensor: shape=(303, 5), dtype=float64, numpy=
array([[ 63. , 150. , 145. , 233. , 2.3],
[ 67. , 108. , 160. , 286. , 1.5],
[ 67. , 129. , 120. , 229. , 2.6],
...,
[ 65. , 127. , 135. , 254. , 2.8],
[ 48. , 150. , 130. , 256. , 0. ],
[ 63. , 154. , 150. , 407. , 4. ]])>
d negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.679903 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.681941 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.683916 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.685875 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.687852 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.689776 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.691767 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.693745 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.695704 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.733646 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.735847 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.737865 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.739866 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.741853 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.743844 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.745817 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.747788 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.749827 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.752365 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.754754 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
I0000 00:00:1723791581.757175 189972 cuda_executor.cc:1015] successful NUMA node read from SysFS had negative value (-1), but there must be at least one NUMA node, so returning NUMA node zero. See more at https://github.com/torvalds/linux/blob/v6.0/Documentation/ABI/testing/sysfs-bus-pci#L344-L355
In general, if an object can be converted to a tensor with tf.convert_to_tensor it can be passed anywhere you can pass a tf.Tensor.
With Model.fit
A DataFrame, interpreted as a single tensor, can be used directly as an argument to the Model.fit method.
Below is an example of training a model on the numeric features of the dataset.
The first step is to normalize the input ranges. Use a tf.keras.layers.Normalization layer for that.
To set the layer's mean and standard-deviation before running it be sure to call the Normalization.adapt method:
normalizer = tf.keras.layers.Normalization(axis=-1)
normalizer.adapt(np.array(numeric_features))
Call the layer on the first three rows of the DataFrame to visualize an example of the output from this layer:
normalizer(numeric_features.iloc[:3])
<tf.Tensor: shape=(3, 5), dtype=float32, numpy=
array([[ 0.93384 , 0.03480783, 0.74578166, -0.26008663, 1.0680454 ],
[ 1.3782114 , -1.7806157 , 1.5923294 , 0.75738776, 0.3802287 ],
[ 1.3782114 , -0.87290394, -0.6651312 , -0.33687717, 1.3259766 ]],
dtype=float32)>
Use the normalization layer as the first layer of a simple model:
def get_basic_model():
model = tf.keras.Sequential([
normalizer,
tf.keras.layers.Dense(10, activation='relu'),
tf.keras.layers.Dense(10, activation='relu'),
tf.keras.layers.Dense(1)
])
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
return model
When you pass the DataFrame as the x argument to Model.fit, Keras treats the DataFrame as it would a NumPy array:
model = get_basic_model()
model.fit(numeric_features, target, epochs=15, batch_size=BATCH_SIZE)
Epoch 1/15 WARNING: All log messages before absl::InitializeLog() is called are written to STDERR I0000 00:00:1723791583.615441 190138 service.cc:146] XLA service 0x7f944c00a090 initialized for platform CUDA (this does not guarantee that XLA will be used). Devices: I0000 00:00:1723791583.615472 190138 service.cc:154] StreamExecutor device (0): Tesla T4, Compute Capability 7.5 I0000 00:00:1723791583.615476 190138 service.cc:154] StreamExecutor device (1): Tesla T4, Compute Capability 7.5 I0000 00:00:1723791583.615480 190138 service.cc:154] StreamExecutor device (2): Tesla T4, Compute Capability 7.5 I0000 00:00:1723791583.615482 190138 service.cc:154] StreamExecutor device (3): Tesla T4, Compute Capability 7.5 115/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.6599 - loss: 0.6927 I0000 00:00:1723791584.314363 190138 device_compiler.h:188] Compiled cluster using XLA! This line is logged at most once for the lifetime of the process. 152/152 ━━━━━━━━━━━━━━━━━━━━ 2s 4ms/step - accuracy: 0.6782 - loss: 0.6784 Epoch 2/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7131 - loss: 0.5535 Epoch 3/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7406 - loss: 0.4835 Epoch 4/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7738 - loss: 0.4418 Epoch 5/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7853 - loss: 0.4522 Epoch 6/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.8086 - loss: 0.4259 Epoch 7/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7701 - loss: 0.4674 Epoch 8/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7587 - loss: 0.4375 Epoch 9/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.8189 - loss: 0.3755 Epoch 10/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7763 - loss: 0.4552 Epoch 11/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.8058 - loss: 0.4008 Epoch 12/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.8209 - loss: 0.4230 Epoch 13/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.8197 - loss: 0.4230 Epoch 14/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.8304 - loss: 0.4119 Epoch 15/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.8232 - loss: 0.3414 <keras.src.callbacks.history.History at 0x7f9604097970>
With tf.data
If you want to apply tf.data transformations to a DataFrame of a uniform dtype, the Dataset.from_tensor_slices method will create a dataset that iterates over the rows of the DataFrame. Each row is initially a vector of values. To train a model, you need (inputs, labels) pairs, so pass (features, labels) and Dataset.from_tensor_slices will return the needed pairs of slices:
numeric_dataset = tf.data.Dataset.from_tensor_slices((numeric_features, target))
for row in numeric_dataset.take(3):
print(row)
(<tf.Tensor: shape=(5,), dtype=float64, numpy=array([ 63. , 150. , 145. , 233. , 2.3])>, <tf.Tensor: shape=(), dtype=int64, numpy=0>) (<tf.Tensor: shape=(5,), dtype=float64, numpy=array([ 67. , 108. , 160. , 286. , 1.5])>, <tf.Tensor: shape=(), dtype=int64, numpy=1>) (<tf.Tensor: shape=(5,), dtype=float64, numpy=array([ 67. , 129. , 120. , 229. , 2.6])>, <tf.Tensor: shape=(), dtype=int64, numpy=0>)
numeric_batches = numeric_dataset.shuffle(1000).batch(BATCH_SIZE)
model = get_basic_model()
model.fit(numeric_batches, epochs=15)
Epoch 1/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 2s 4ms/step - accuracy: 0.6986 - loss: 0.6073 Epoch 2/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7271 - loss: 0.5204 Epoch 3/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7176 - loss: 0.4899 Epoch 4/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7307 - loss: 0.4668 Epoch 5/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7695 - loss: 0.4536 Epoch 6/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7448 - loss: 0.4346 Epoch 7/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7832 - loss: 0.4221 Epoch 8/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7845 - loss: 0.4367 Epoch 9/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7622 - loss: 0.4458 Epoch 10/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7874 - loss: 0.4647 Epoch 11/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7821 - loss: 0.4104 Epoch 12/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7588 - loss: 0.4802 Epoch 13/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.7568 - loss: 0.4469 Epoch 14/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.8218 - loss: 0.4112 Epoch 15/15 152/152 ━━━━━━━━━━━━━━━━━━━━ 0s 1ms/step - accuracy: 0.8091 - loss: 0.3851 <keras.src.callbacks.history.History at 0x7f95f0333be0>
A DataFrame as a dictionary
When you start dealing with heterogeneous data, it is no longer possible to treat the DataFrame as if it were a single array. TensorFlow tensors require that all elements have the same dtype.
So, in this case, you need to start treating it as a dictionary of columns, where each column has a uniform dtype. A DataFrame is a lot like a dictionary of arrays, so typically all you need to do is cast the DataFrame to a Python dict. Many important TensorFlow APIs support (nested-)dictionaries of arrays as inputs.
tf.data input pipelines handle this quite well. All tf.data operations handle dictionaries and tuples automatically. So, to make a dataset of dictionary-examples from a DataFrame, just cast it to a dict before slicing it with Dataset.from_tensor_slices:
numeric_features_dict = {key: value.to_numpy()[:, tf.newaxis] for key, value in dict(numeric_features).items()}
target_array = target.to_numpy()[:, tf.newaxis]
numeric_dict_ds = tf.data.Dataset.from_tensor_slices((numeric_features_dict , target_array))
len(numeric_features_dict)
5
Here are the first three examples from that dataset:
for row in numeric_dict_ds.take(3):
print(row)
({'age': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([63])>, 'thalach': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([150])>, 'trestbps': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([145])>, 'chol': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([233])>, 'oldpeak': <tf.Tensor: shape=(1,), dtype=float64, numpy=array([2.3])>}, <tf.Tensor: shape=(1,), dtype=int64, numpy=array([0])>)
({'age': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([67])>, 'thalach': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([108])>, 'trestbps': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([160])>, 'chol': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([286])>, 'oldpeak': <tf.Tensor: shape=(1,), dtype=float64, numpy=array([1.5])>}, <tf.Tensor: shape=(1,), dtype=int64, numpy=array([1])>)
({'age': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([67])>, 'thalach': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([129])>, 'trestbps': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([120])>, 'chol': <tf.Tensor: shape=(1,), dtype=int64, numpy=array([229])>, 'oldpeak': <tf.Tensor: shape=(1,), dtype=float64, numpy=array([2.6])>}, <tf.Tensor: shape=(1,), dtype=int64, numpy=array([0])>)
Typically, Keras models and layers expect a single input tensor, but these classes can accept and return nested structures of dictionaries, tuples and tensors. These structures are known as "nests" (refer to the tf.nest module for details).
There are two equivalent ways you can write a Keras model that accepts a dictionary as input.
1. The Model-subclass style
You write a subclass of tf.keras.Model (or tf.keras.Layer). You directly handle the inputs, and create the outputs:
class MyModel(tf.keras.Model):
def __init__(self):
# Create all the internal layers in init.
super().__init__()
self.normalizer = tf.keras.layers.Normalization(axis=-1)
self.seq = tf.keras.Sequential([
self.normalizer,
tf.keras.layers.Dense(10, activation='relu'),
tf.keras.layers.Dense(10, activation='relu'),
tf.keras.layers.Dense(1)
])
self.concat = tf.keras.layers.Concatenate(axis=1)
def _stack(self, input_dict):
values = []
for key, value in sorted(input_dict.items()):
values.append(value)
return self.concat(values)
def adapt(self, inputs):
# Stack the inputs and `adapt` the normalization layer.
inputs = self._stack(inputs)
self.normalizer.adapt(inputs)
def call(self, inputs):
# Stack the inputs
inputs = self._stack(inputs)
# Run them through all the layers.
result = self.seq(inputs)
return result
model = MyModel()
model.adapt(numeric_features_dict)
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'],
run_eagerly=True)
This model can accept either a dictionary of columns or a dataset of dictionary-elements for training:
model.fit(numeric_features_dict, target_array, epochs=5, batch_size=BATCH_SIZE)
Epoch 1/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 8s 44ms/step - accuracy: 0.6925 - loss: 0.6956 Epoch 2/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 43ms/step - accuracy: 0.7367 - loss: 0.5419 Epoch 3/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 43ms/step - accuracy: 0.7023 - loss: 0.4857 Epoch 4/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7300 - loss: 0.4832 Epoch 5/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7624 - loss: 0.4132 <keras.src.callbacks.history.History at 0x7f95b8f49520>
numeric_dict_batches = numeric_dict_ds.shuffle(SHUFFLE_BUFFER).batch(BATCH_SIZE)
model.fit(numeric_dict_batches, epochs=5)
Epoch 1/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7488 - loss: 0.4628 Epoch 2/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7816 - loss: 0.4485 Epoch 3/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7621 - loss: 0.4226 Epoch 4/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7848 - loss: 0.4282 Epoch 5/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7320 - loss: 0.4743 <keras.src.callbacks.history.History at 0x7f95b8f3af40>
Here are the predictions for the first three examples:
model.predict(dict(numeric_features.iloc[:3]))
1/1 ━━━━━━━━━━━━━━━━━━━━ 0s 52ms/step
array([[0.07420629],
[0.00349799],
[0.28239426]], dtype=float32)
2. The Keras functional style
inputs = {}
for name, column in numeric_features.items():
inputs[name] = tf.keras.Input(
shape=(1,), name=name, dtype=tf.float32)
inputs
{'age': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=age>,
'thalach': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=thalach>,
'trestbps': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=trestbps>,
'chol': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=chol>,
'oldpeak': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=oldpeak>}
xs = [value for key, value in sorted(inputs.items())]
concat = tf.keras.layers.Concatenate(axis=1)
x = concat(xs)
normalizer = tf.keras.layers.Normalization(axis=-1)
normalizer.adapt(np.concatenate([value for key, value in sorted(numeric_features_dict.items())], axis=1))
x = normalizer(x)
x = tf.keras.layers.Dense(10, activation='relu')(x)
x = tf.keras.layers.Dense(10, activation='relu')(x)
x = tf.keras.layers.Dense(1)(x)
model = tf.keras.Model(inputs, x)
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'],
run_eagerly=True)
tf.keras.utils.plot_model(model, rankdir="LR", show_shapes=True, show_layer_names=True)

You can train the functional model the same way as the model subclass:
model.fit(numeric_features_dict, target, epochs=5, batch_size=BATCH_SIZE)
Epoch 1/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 43ms/step - accuracy: 0.7544 - loss: 0.7001 Epoch 2/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7164 - loss: 0.5951 Epoch 3/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7140 - loss: 0.5189 Epoch 4/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7545 - loss: 0.4637 Epoch 5/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7454 - loss: 0.4758 <keras.src.callbacks.history.History at 0x7f95b8eaeee0>
numeric_dict_batches = numeric_dict_ds.shuffle(SHUFFLE_BUFFER).batch(BATCH_SIZE)
model.fit(numeric_dict_batches, epochs=5)
Epoch 1/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 43ms/step - accuracy: 0.7140 - loss: 0.4563 Epoch 2/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 43ms/step - accuracy: 0.7363 - loss: 0.4123 Epoch 3/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7138 - loss: 0.4769 Epoch 4/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7535 - loss: 0.3938 Epoch 5/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 7s 44ms/step - accuracy: 0.7452 - loss: 0.4386 <keras.src.callbacks.history.History at 0x7f960804ed00>
Full example
If you're passing a heterogeneous DataFrame to Keras, each column may need unique preprocessing. You could do this preprocessing directly in the DataFrame, but for a model to work correctly, inputs always need to be preprocessed the same way. So, the best approach is to build the preprocessing into the model. Keras preprocessing layers cover many common tasks.
Build the preprocessing head
In this dataset some of the "integer" features in the raw data are actually Categorical indices. These indices are not really ordered numeric values (refer to the the dataset description for details). Because these are unordered they are inappropriate to feed directly to the model; the model would interpret them as being ordered. To use these inputs you'll need to encode them, either as one-hot vectors or embedding vectors. The same applies to string-categorical features.
Binary features on the other hand do not generally need to be encoded or normalized.
Start by by creating a list of the features that fall into each group:
binary_feature_names = ['sex', 'fbs', 'exang']
categorical_feature_names = ['cp', 'restecg', 'slope', 'thal', 'ca']
The next step is to build a preprocessing model that will apply appropriate preprocessing to each input and concatenate the results.
This section uses the Keras Functional API to implement the preprocessing. You start by creating one tf.keras.Input for each column of the dataframe:
inputs = {}
for name, column in df.items():
if type(column[0]) == str:
dtype = tf.string
elif (name in categorical_feature_names or
name in binary_feature_names):
dtype = tf.int64
else:
dtype = tf.float32
inputs[name] = tf.keras.Input(shape=(1,), name=name, dtype=dtype)
inputs
{'age': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=age>,
'sex': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=sex>,
'cp': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=cp>,
'trestbps': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=trestbps>,
'chol': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=chol>,
'fbs': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=fbs>,
'restecg': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=restecg>,
'thalach': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=thalach>,
'exang': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=exang>,
'oldpeak': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=oldpeak>,
'slope': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=slope>,
'ca': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=ca>,
'thal': <KerasTensor shape=(None, 1), dtype=string, sparse=False, name=thal>}
For each input you'll apply some transformations using Keras layers and TensorFlow ops. Each feature starts as a batch of scalars (shape=(batch,)). The output for each should be a batch of tf.float32 vectors (shape=(batch, n)). The last step will concatenate all those vectors together.
Binary inputs
Since the binary inputs don't need any preprocessing, just add the vector axis, cast them to float32 and add them to the list of preprocessed inputs:
preprocessed = []
for name in binary_feature_names:
inp = inputs[name]
preprocessed.append(inp)
preprocessed
[<KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=sex>, <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=fbs>, <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=exang>]
Numeric inputs
Like in the earlier section you'll want to run these numeric inputs through a tf.keras.layers.Normalization layer before using them. The difference is that this time they're input as a dict. The code below collects the numeric features from the DataFrame, stacks them together and passes those to the Normalization.adapt method.
normalizer = tf.keras.layers.Normalization(axis=-1)
normalizer.adapt(np.concatenate([value for key, value in sorted(numeric_features_dict.items())], axis=1))
The code below stacks the numeric features and runs them through the normalization layer.
numeric_inputs = []
for name in numeric_feature_names:
numeric_inputs.append(inputs[name])
numeric_inputs = tf.keras.layers.Concatenate(axis=-1)(numeric_inputs)
numeric_normalized = normalizer(numeric_inputs)
preprocessed.append(numeric_normalized)
preprocessed
[<KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=sex>, <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=fbs>, <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=exang>, <KerasTensor shape=(None, 5), dtype=float32, sparse=False, name=keras_tensor_22>]
Categorical features
To use categorical features you'll first need to encode them into either binary vectors or embeddings. Since these features only contain a small number of categories, convert the inputs directly to one-hot vectors using the output_mode='one_hot' option, supported by both the tf.keras.layers.StringLookup and tf.keras.layers.IntegerLookup layers.
Here is an example of how these layers work:
vocab = ['a','b','c']
lookup = tf.keras.layers.StringLookup(vocabulary=vocab, output_mode='one_hot')
lookup(['c','a','a','b','zzz'])
<tf.Tensor: shape=(5, 4), dtype=int64, numpy=
array([[0, 0, 0, 1],
[0, 1, 0, 0],
[0, 1, 0, 0],
[0, 0, 1, 0],
[1, 0, 0, 0]])>
vocab = [1,4,7,99]
lookup = tf.keras.layers.IntegerLookup(vocabulary=vocab, output_mode='one_hot')
lookup([-1,4,1])
<tf.Tensor: shape=(3, 5), dtype=int64, numpy=
array([[1, 0, 0, 0, 0],
[0, 0, 1, 0, 0],
[0, 1, 0, 0, 0]])>
To determine the vocabulary for each input, create a layer to convert that vocabulary to a one-hot vector:
for name in categorical_feature_names:
vocab = sorted(set(df[name]))
print(f'name: {name}')
print(f'vocab: {vocab}\n')
if type(vocab[0]) is str:
lookup = tf.keras.layers.StringLookup(vocabulary=vocab, output_mode='one_hot')
else:
lookup = tf.keras.layers.IntegerLookup(vocabulary=vocab, output_mode='one_hot')
x = inputs[name]
x = lookup(x)
preprocessed.append(x)
name: cp vocab: [0, 1, 2, 3, 4] name: restecg vocab: [0, 1, 2] name: slope vocab: [1, 2, 3] name: thal vocab: ['1', '2', 'fixed', 'normal', 'reversible'] name: ca vocab: [0, 1, 2, 3]
Assemble the preprocessing head
At this point preprocessed is just a Python list of all the preprocessing results, each result has a shape of (batch_size, depth):
preprocessed
[<KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=sex>, <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=fbs>, <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=exang>, <KerasTensor shape=(None, 5), dtype=float32, sparse=False, name=keras_tensor_22>, <KerasTensor shape=(None, 6), dtype=float32, sparse=False, name=keras_tensor_23>, <KerasTensor shape=(None, 4), dtype=float32, sparse=False, name=keras_tensor_24>, <KerasTensor shape=(None, 4), dtype=float32, sparse=False, name=keras_tensor_25>, <KerasTensor shape=(None, 6), dtype=float32, sparse=False, name=keras_tensor_26>, <KerasTensor shape=(None, 5), dtype=float32, sparse=False, name=keras_tensor_27>]
Concatenate all the preprocessed features along the depth axis, so each dictionary-example is converted into a single vector. The vector contains categorical features, numeric features, and categorical one-hot features:
preprocessed_result = tf.keras.layers.Concatenate(axis=1)(preprocessed)
preprocessed_result
<KerasTensor shape=(None, 33), dtype=float32, sparse=False, name=keras_tensor_28>
Now create a model out of that calculation so it can be reused:
preprocessor = tf.keras.Model(inputs, preprocessed_result)
tf.keras.utils.plot_model(preprocessor, rankdir="LR", show_shapes=True, show_layer_names=True)
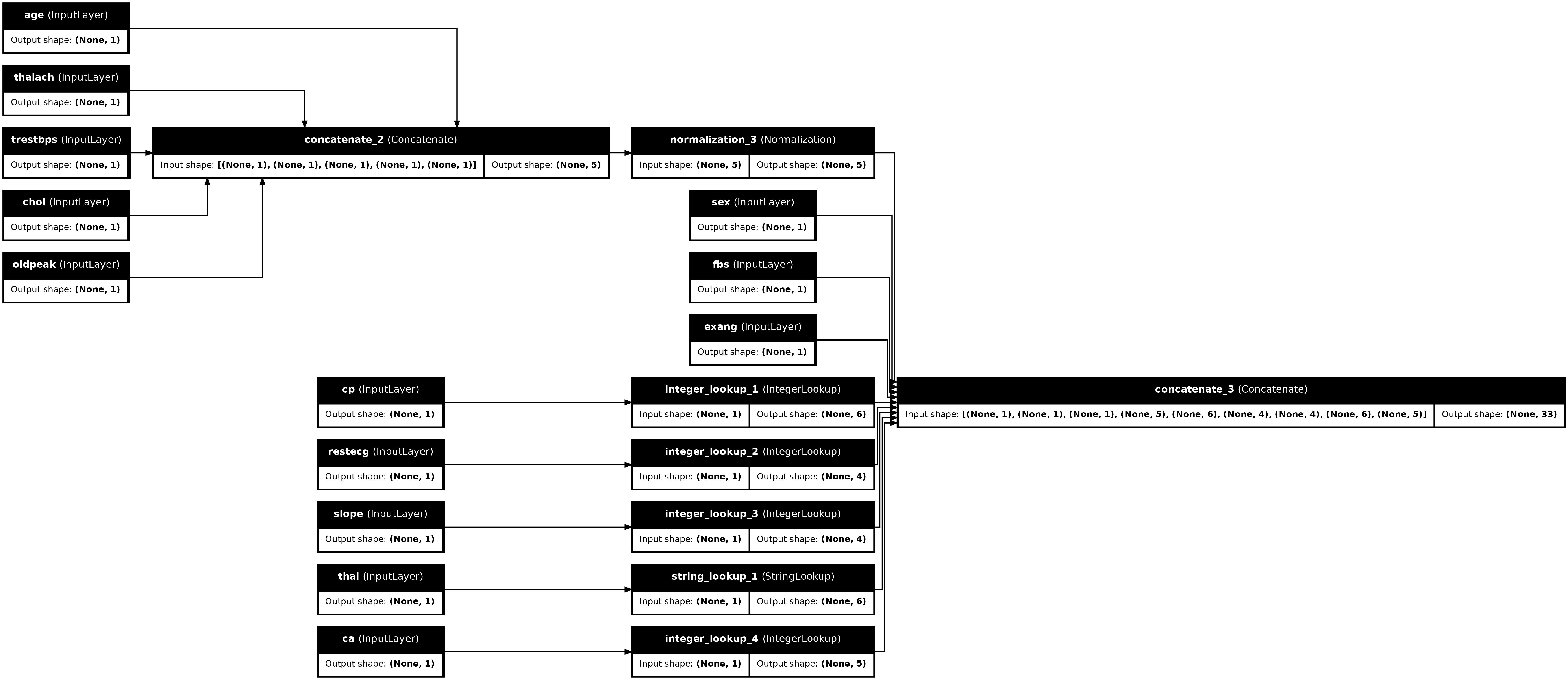
To test the preprocessor, use the DataFrame.iloc accessor to slice the first example from the DataFrame. Then convert it to a dictionary and pass the dictionary to the preprocessor. The result is a single vector containing the binary features, normalized numeric features and the one-hot categorical features, in that order:
preprocessor(dict(df.iloc[:1]))
<tf.Tensor: shape=(1, 33), dtype=float32, numpy=
array([[ 1. , 1. , 0. , 0.93384 , -1.8534899,
123.75736 , 3.6224306, -7.3077087, 0. , 0. ,
1. , 0. , 0. , 0. , 0. ,
0. , 0. , 1. , 0. , 0. ,
0. , 1. , 0. , 0. , 0. ,
1. , 0. , 0. , 0. , 1. ,
0. , 0. , 0. ]], dtype=float32)>
Create and train a model
Now build the main body of the model. Use the same configuration as in the previous example: A couple of Dense rectified-linear layers and a Dense(1) output layer for the classification.
body = tf.keras.Sequential([
tf.keras.layers.Dense(10, activation='relu'),
tf.keras.layers.Dense(10, activation='relu'),
tf.keras.layers.Dense(1)
])
Now put the two pieces together using the Keras functional API.
inputs
{'age': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=age>,
'sex': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=sex>,
'cp': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=cp>,
'trestbps': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=trestbps>,
'chol': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=chol>,
'fbs': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=fbs>,
'restecg': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=restecg>,
'thalach': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=thalach>,
'exang': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=exang>,
'oldpeak': <KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=oldpeak>,
'slope': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=slope>,
'ca': <KerasTensor shape=(None, 1), dtype=int64, sparse=False, name=ca>,
'thal': <KerasTensor shape=(None, 1), dtype=string, sparse=False, name=thal>}
x = preprocessor(inputs)
x
<KerasTensor shape=(None, 33), dtype=float32, sparse=False, name=keras_tensor_29>
result = body(x)
result
<KerasTensor shape=(None, 1), dtype=float32, sparse=False, name=keras_tensor_34>
model = tf.keras.Model(inputs, result)
model.compile(optimizer='adam',
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=['accuracy'])
tf.keras.utils.plot_model(model, show_shapes=True, show_layer_names=True)
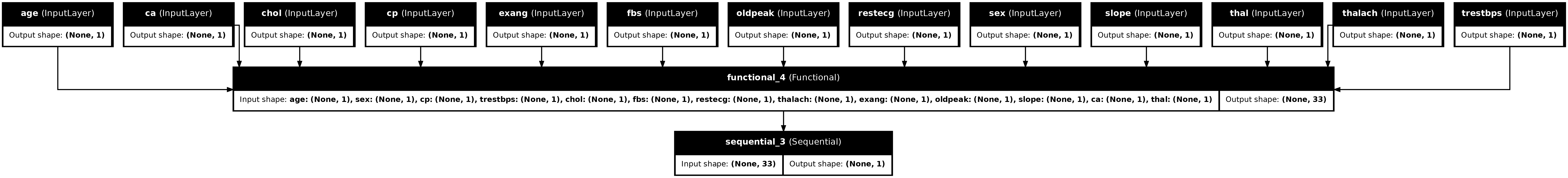
This model expects a dictionary of inputs. The simplest way to pass it the data is to convert the DataFrame to a dict and pass that dict as the x argument to Model.fit:
history = model.fit(dict(df), target, epochs=5, batch_size=BATCH_SIZE)
Epoch 1/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 2s 4ms/step - accuracy: 0.4448 - loss: 2.5713 Epoch 2/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 1s 4ms/step - accuracy: 0.6752 - loss: 0.6143 Epoch 3/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 1s 4ms/step - accuracy: 0.7503 - loss: 0.5409 Epoch 4/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 1s 4ms/step - accuracy: 0.7133 - loss: 0.5542 Epoch 5/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 1s 4ms/step - accuracy: 0.6721 - loss: 0.5666
Using tf.data works as well:
ds = tf.data.Dataset.from_tensor_slices((
dict(df),
target
))
ds = ds.batch(BATCH_SIZE)
import pprint
for x, y in ds.take(1):
pprint.pprint(x)
print()
print(y)
{'age': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([63, 67])>,
'ca': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([0, 3])>,
'chol': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([233, 286])>,
'cp': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([1, 4])>,
'exang': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([0, 1])>,
'fbs': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([1, 0])>,
'oldpeak': <tf.Tensor: shape=(2,), dtype=float64, numpy=array([2.3, 1.5])>,
'restecg': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([2, 2])>,
'sex': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([1, 1])>,
'slope': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([3, 2])>,
'thal': <tf.Tensor: shape=(2,), dtype=string, numpy=array([b'fixed', b'normal'], dtype=object)>,
'thalach': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([150, 108])>,
'trestbps': <tf.Tensor: shape=(2,), dtype=int64, numpy=array([145, 160])>}
tf.Tensor([0 1], shape=(2,), dtype=int64)
history = model.fit(ds, epochs=5)
Epoch 1/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 1s 4ms/step - accuracy: 0.7511 - loss: 0.4749 Epoch 2/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 1s 4ms/step - accuracy: 0.7511 - loss: 0.4700 Epoch 3/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 1s 4ms/step - accuracy: 0.7626 - loss: 0.4490 Epoch 4/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 1s 4ms/step - accuracy: 0.7543 - loss: 0.4288 Epoch 5/5 152/152 ━━━━━━━━━━━━━━━━━━━━ 1s 4ms/step - accuracy: 0.7537 - loss: 0.4099
