- Description:
The CBIS-DDSM (Curated Breast Imaging Subset of DDSM) is an updated and standardized version of the Digital Database for Screening Mammography (DDSM). The DDSM is a database of 2,620 scanned film mammography studies. It contains normal, benign, and malignant cases with verified pathology information.
The default config is made of patches extracted from the original mammograms, following the description from (http://arxiv.org/abs/1708.09427), in order to frame the task to solve in a traditional image classification setting.
Homepage: https://wiki.cancerimagingarchive.net/display/Public/CBIS-DDSM
Source code:
tfds.image_classification.CuratedBreastImagingDDSMVersions:
2.0.1: New split API (https://tensorflow.org/datasets/splits)3.0.0(default): Better cropping sampling (https://github.com/tensorflow/datasets/pull/2502)
Manual download instructions: This dataset requires you to download the source data manually into
download_config.manual_dir(defaults to~/tensorflow_datasets/downloads/manual/):
You can download the images from https://wiki.cancerimagingarchive.net/display/Public/CBIS-DDSM
Because special software and libraries are needed to download and read the images contained in the dataset, TFDS assumes that the user has downloaded the original DCIM files and converted them to PNG.
The following commands (or equivalent) should be used to generate the PNG files, in order to guarantee reproducible results:
find $DATASET_DCIM_DIR -name '*.dcm' | \
xargs -n1 -P8 -I{} bash -c 'f={}; dcmj2pnm $f | convert - ${f/.dcm/.png}'
Resulting images should be put in manual_dir, like:
<manual_dir>/Mass-Training_P_01981_RIGHT_MLO_1/1.3.6.../000000.png.
Auto-cached (documentation): No
Supervised keys (See
as_superviseddoc):NoneCitation:
@misc{CBIS_DDSM_Citation,
doi = {10.7937/k9/tcia.2016.7o02s9cy},
url = {https://wiki.cancerimagingarchive.net/x/lZNXAQ},
author = {Sawyer-Lee, Rebecca and Gimenez, Francisco and Hoogi, Assaf and Rubin, Daniel},
title = {Curated Breast Imaging Subset of DDSM},
publisher = {The Cancer Imaging Archive},
year = {2016},
}
@article{TCIA_Citation,
author = {
K. Clark and B. Vendt and K. Smith and J. Freymann and J. Kirby and
P. Koppel and S. Moore and S. Phillips and D. Maffitt and M. Pringle and
L. Tarbox and F. Prior
},
title = { {The Cancer Imaging Archive (TCIA): Maintaining and Operating a
Public Information Repository} },
journal = {Journal of Digital Imaging},
volume = {26},
month = {December},
year = {2013},
pages = {1045-1057},
}
@article{DBLP:journals/corr/abs-1708-09427,
author = {Li Shen},
title = {End-to-end Training for Whole Image Breast Cancer Diagnosis using
An All Convolutional Design},
journal = {CoRR},
volume = {abs/1708.09427},
year = {2017},
url = {http://arxiv.org/abs/1708.09427},
archivePrefix = {arXiv},
eprint = {1708.09427},
timestamp = {Mon, 13 Aug 2018 16:48:35 +0200},
biburl = {https://dblp.org/rec/bib/journals/corr/abs-1708-09427},
bibsource = {dblp computer science bibliography, https://dblp.org}
}
curated_breast_imaging_ddsm/patches (default config)
Config description: Patches containing both calsification and mass cases, plus pathces with no abnormalities. Designed as a traditional 5-class classification task.
Download size:
2.01 MiBDataset size:
801.46 MiBSplits:
| Split | Examples |
|---|---|
'test' |
9,770 |
'train' |
49,780 |
'validation' |
5,580 |
- Feature structure:
FeaturesDict({
'id': Text(shape=(), dtype=string),
'image': Image(shape=(None, None, 1), dtype=uint8),
'label': ClassLabel(shape=(), dtype=int64, num_classes=5),
})
- Feature documentation:
| Feature | Class | Shape | Dtype | Description |
|---|---|---|---|---|
| FeaturesDict | ||||
| id | Text | string | ||
| image | Image | (None, None, 1) | uint8 | |
| label | ClassLabel | int64 |
- Figure (tfds.show_examples):
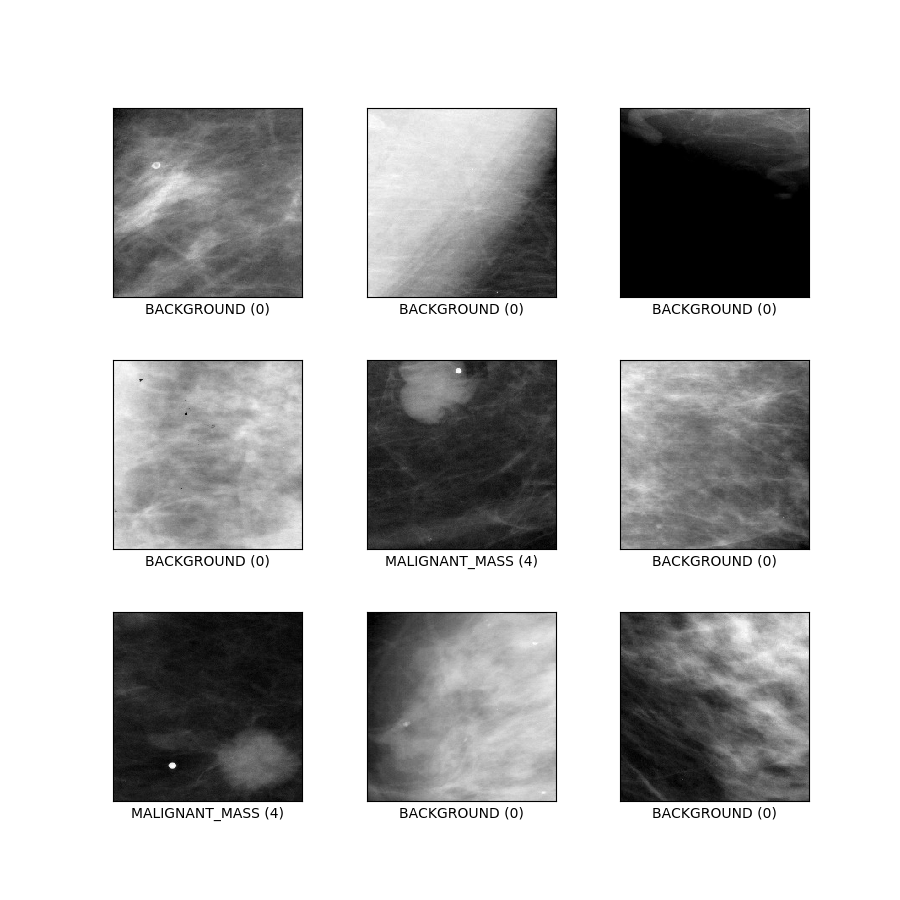
- Examples (tfds.as_dataframe):
curated_breast_imaging_ddsm/original-calc
Config description: Original images of the calcification cases compressed in lossless PNG.
Download size:
1.06 MiBDataset size:
4.42 GiBSplits:
| Split | Examples |
|---|---|
'test' |
284 |
'train' |
1,227 |
- Feature structure:
FeaturesDict({
'abnormalities': Sequence({
'assessment': ClassLabel(shape=(), dtype=int64, num_classes=6),
'calc_distribution': ClassLabel(shape=(), dtype=int64, num_classes=10),
'calc_type': ClassLabel(shape=(), dtype=int64, num_classes=48),
'id': int32,
'mask': Image(shape=(None, None, 1), dtype=uint8),
'pathology': ClassLabel(shape=(), dtype=int64, num_classes=3),
'subtlety': ClassLabel(shape=(), dtype=int64, num_classes=6),
}),
'breast': ClassLabel(shape=(), dtype=int64, num_classes=2),
'id': Text(shape=(), dtype=string),
'image': Image(shape=(None, None, 1), dtype=uint8),
'patient': Text(shape=(), dtype=string),
'view': ClassLabel(shape=(), dtype=int64, num_classes=2),
})
- Feature documentation:
| Feature | Class | Shape | Dtype | Description |
|---|---|---|---|---|
| FeaturesDict | ||||
| abnormalities | Sequence | |||
| abnormalities/assessment | ClassLabel | int64 | ||
| abnormalities/calc_distribution | ClassLabel | int64 | ||
| abnormalities/calc_type | ClassLabel | int64 | ||
| abnormalities/id | Tensor | int32 | ||
| abnormalities/mask | Image | (None, None, 1) | uint8 | |
| abnormalities/pathology | ClassLabel | int64 | ||
| abnormalities/subtlety | ClassLabel | int64 | ||
| breast | ClassLabel | int64 | ||
| id | Text | string | ||
| image | Image | (None, None, 1) | uint8 | |
| patient | Text | string | ||
| view | ClassLabel | int64 |
- Figure (tfds.show_examples):
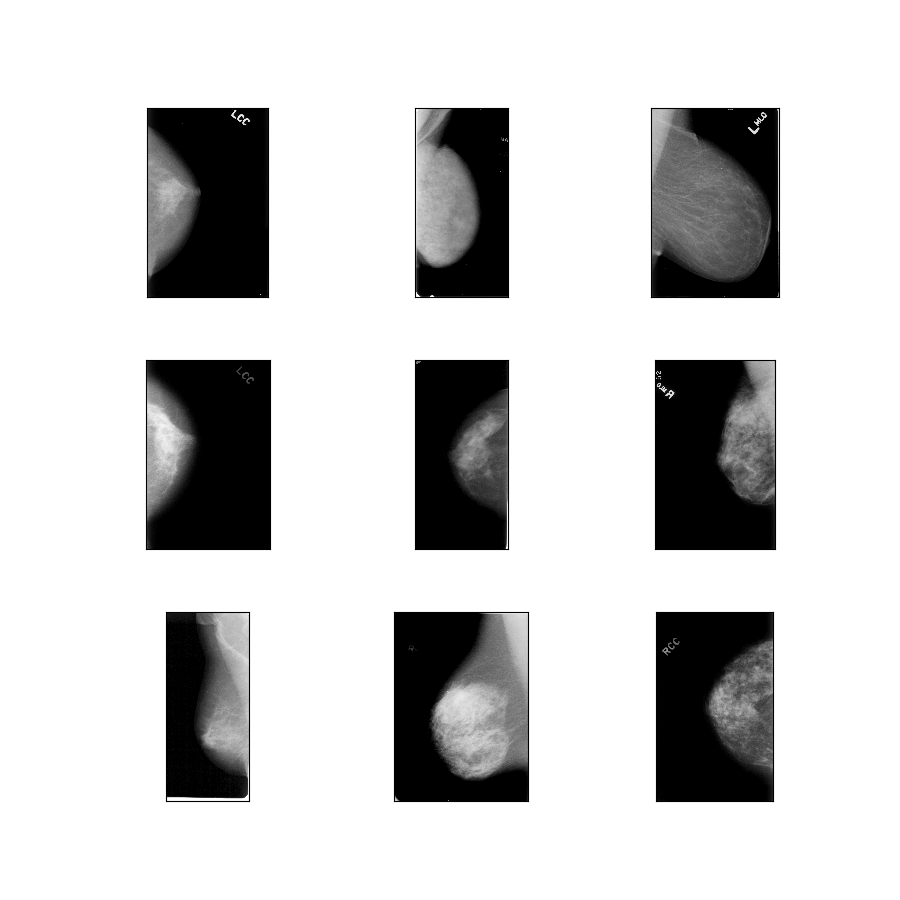
- Examples (tfds.as_dataframe):
curated_breast_imaging_ddsm/original-mass
Config description: Original images of the mass cases compressed in lossless PNG.
Download size:
966.57 KiBDataset size:
4.80 GiBSplits:
| Split | Examples |
|---|---|
'test' |
348 |
'train' |
1,166 |
- Feature structure:
FeaturesDict({
'abnormalities': Sequence({
'assessment': ClassLabel(shape=(), dtype=int64, num_classes=6),
'id': int32,
'mask': Image(shape=(None, None, 1), dtype=uint8),
'mass_margins': ClassLabel(shape=(), dtype=int64, num_classes=20),
'mass_shape': ClassLabel(shape=(), dtype=int64, num_classes=21),
'pathology': ClassLabel(shape=(), dtype=int64, num_classes=3),
'subtlety': ClassLabel(shape=(), dtype=int64, num_classes=6),
}),
'breast': ClassLabel(shape=(), dtype=int64, num_classes=2),
'id': Text(shape=(), dtype=string),
'image': Image(shape=(None, None, 1), dtype=uint8),
'patient': Text(shape=(), dtype=string),
'view': ClassLabel(shape=(), dtype=int64, num_classes=2),
})
- Feature documentation:
| Feature | Class | Shape | Dtype | Description |
|---|---|---|---|---|
| FeaturesDict | ||||
| abnormalities | Sequence | |||
| abnormalities/assessment | ClassLabel | int64 | ||
| abnormalities/id | Tensor | int32 | ||
| abnormalities/mask | Image | (None, None, 1) | uint8 | |
| abnormalities/mass_margins | ClassLabel | int64 | ||
| abnormalities/mass_shape | ClassLabel | int64 | ||
| abnormalities/pathology | ClassLabel | int64 | ||
| abnormalities/subtlety | ClassLabel | int64 | ||
| breast | ClassLabel | int64 | ||
| id | Text | string | ||
| image | Image | (None, None, 1) | uint8 | |
| patient | Text | string | ||
| view | ClassLabel | int64 |
- Figure (tfds.show_examples):
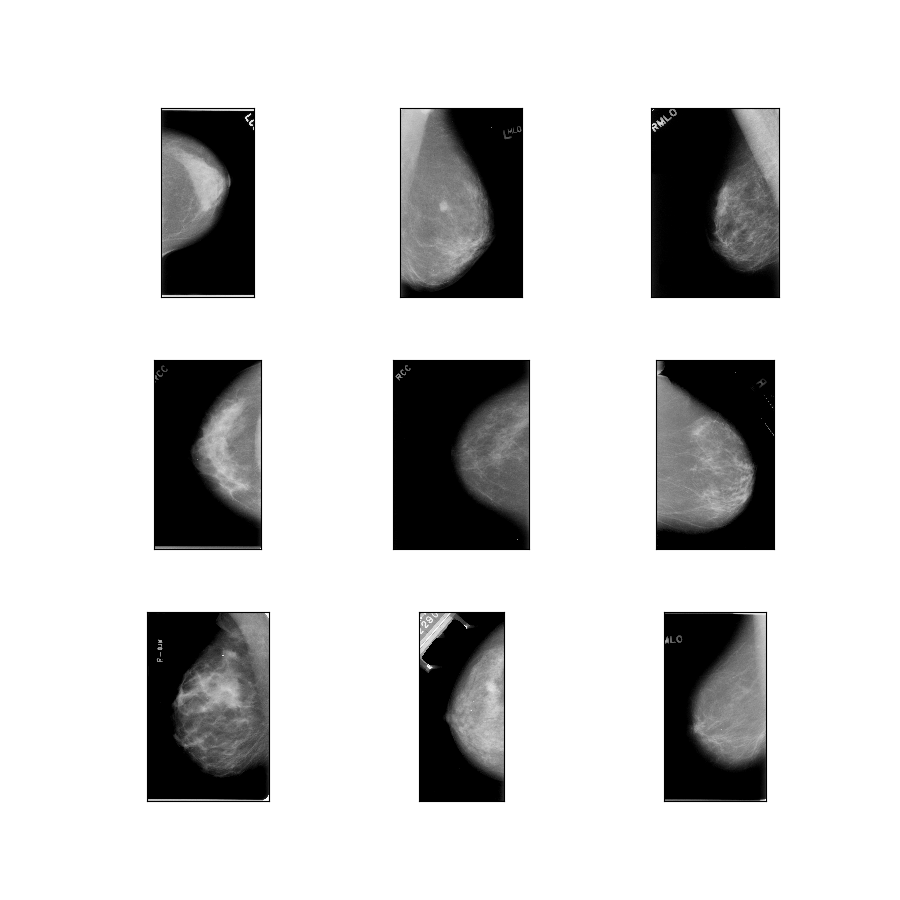
- Examples (tfds.as_dataframe):
